Designing precise all-atom protein structures is a critical challenge in bioengineering as it involves generating both 3D structural data and 1D sequence information to define the placement of side chain atoms. Most approaches currently rely heavily on resolved, experimentally determined structural datasets, which are scarce and biased, limiting exploration of natural protein space. Furthermore, these approaches typically divide the processes of sequence and structure generation, leading to inefficiencies and inadequate integration of multimodal aspects. Removing these obstacles is critical to advancing protein design and enabling advances in drug discovery, molecular engineering, and synthetic biology.
Traditional protein design methods rely on backbone structure prediction or inverse folding models to derive sequences from known structures. These methods are limited by the separation of modalities, as many techniques treat sequence and structure as different tasks, thereby making their integration difficult. A significant reliance on experimentally determined structural data sets excludes a significant number of non-crystallizable proteins and limits generalizability. Switching between structure prediction and sequence generation increases runtime and increases the risk of error propagation. Current tools also face the challenge of adapting large-scale, function-specific protein designs due to rigid structures and limitations in datasets. These limitations limit the scope of existing methods, particularly with regard to the design of proteins that require high multimodal accuracy and variation.
Researchers from UC Berkeley, Genentech, Microsoft Research and New York University introduce PLAID (Protein Latent Induced Diffusion), a generative framework that synthesizes sequence and all-atom protein structures by leveraging latent diffusion. Such a framework leverages the cohesive latent space derived from ESMFold for smooth multimodal integration and reduces the dependence on external prediction models. This approach increases the data distribution by 2–4 orders of magnitude over structural datasets to better cover natural protein diversity. It uses a Diffusion Transformer architecture that is hardware-oriented and can be scaled for conditional multimodal protein generation. Its functional and organism-conditioned samples achieve high structural fidelity and biophysical relevance for a wide range of protein design requirements. These features describe major advances in enabling diverse, scalable, and application-driven protein production.
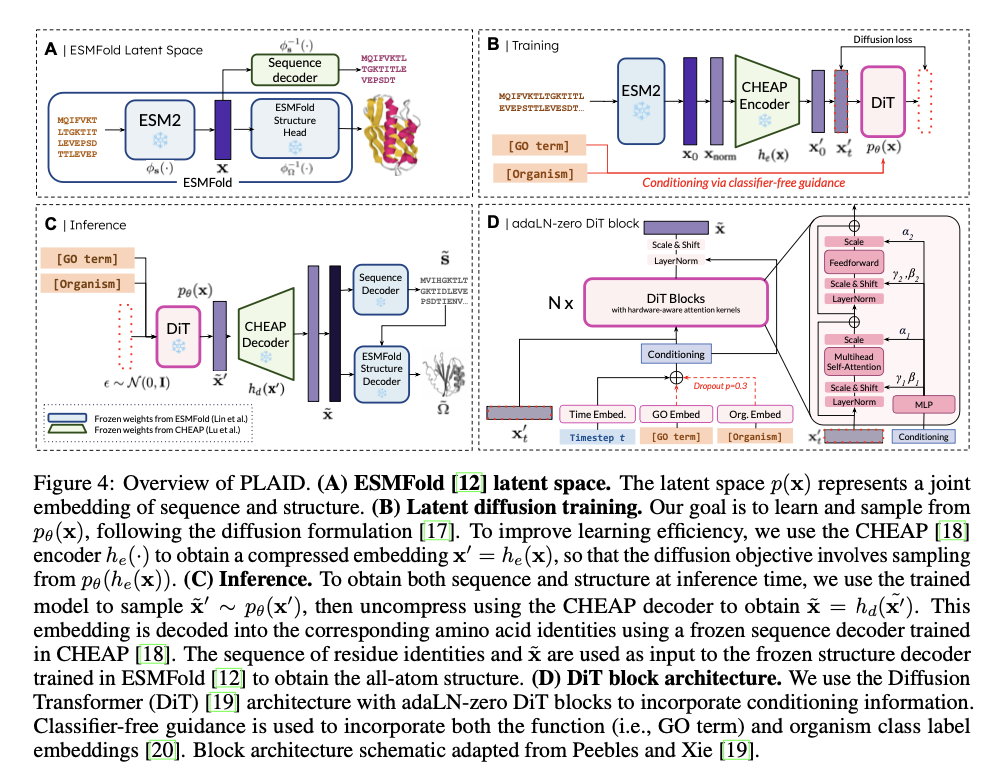
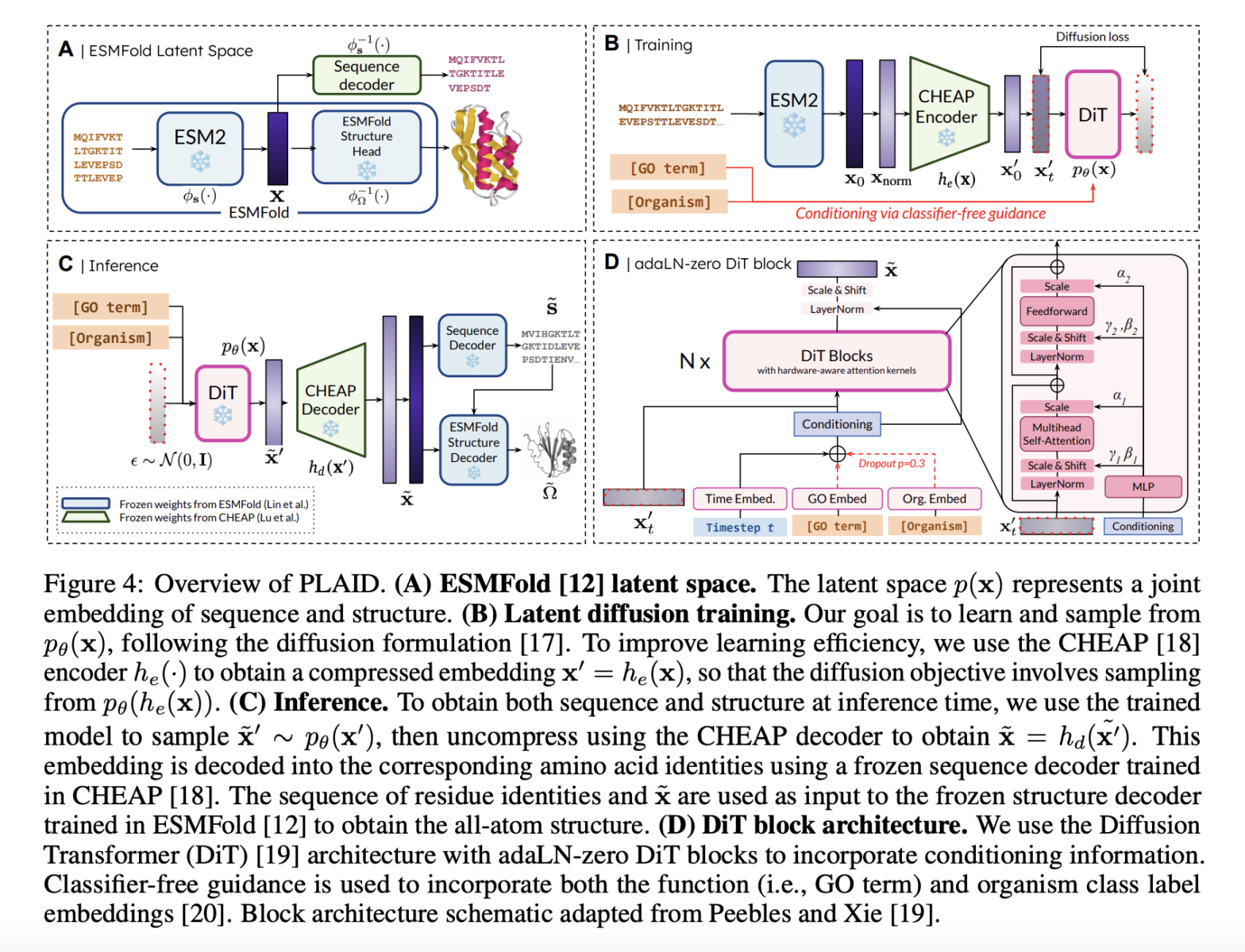
PLAID works by correlating protein sequences with a reduced latent space that is fine-tuned for efficient diffusion and generation. Indirect capture of structural constraints is achieved by using pre-trained ESMFold weights, as there is a wealth of annotated sequence data in databases such as Pfam. A discrete-time diffusion framework systematically improves latent embeddings while integrating classifier-free guidance for functional and organismal conditioning. It includes a CHEAP autoencoder for dimensionality reduction and Diffusion Transformer blocks for noise reduction, making it scalable for long protein sequences. Metrics include cross-modal consistency, self-consistency, and conformance to biophysical distributions of natural proteins, validating the quality and biological relevance of the generated samples.
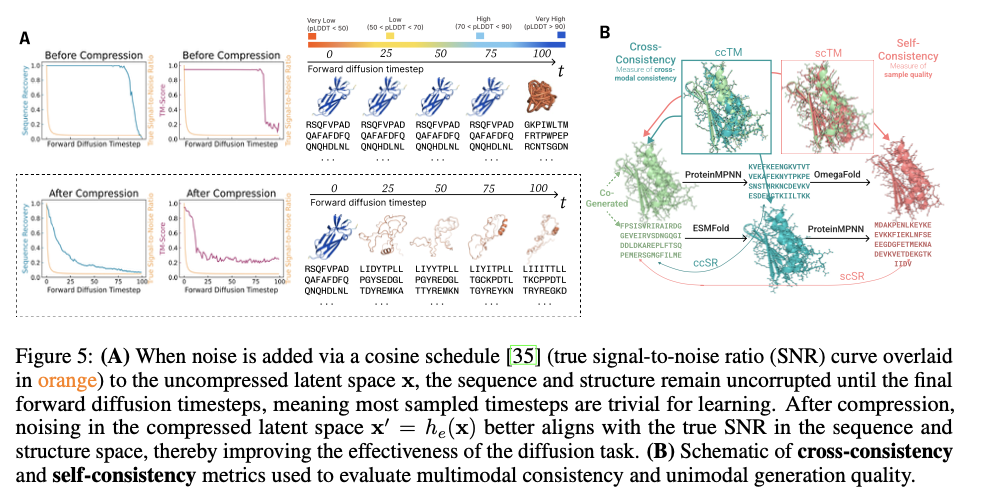
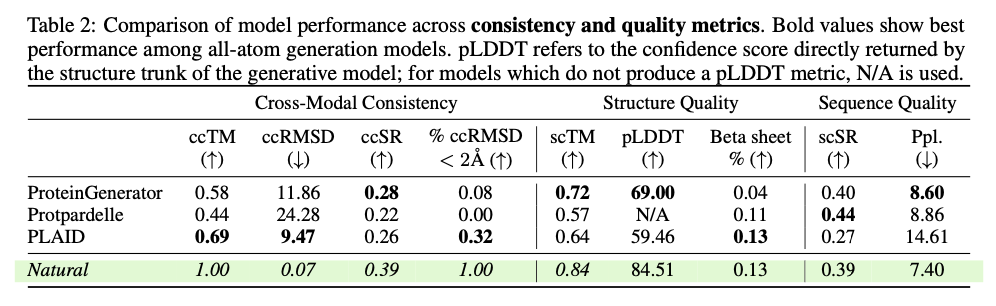
This innovative framework demonstrates significant progress in creating biologically relevant and structurally precise protein designs. PLAID exhibits high cross-modal consistency and therefore shows significant improvements in the alignment of generated sequences and structures as well as improved designability compared to previous approaches. The produced proteins exhibit robust structural fidelity, capture biophysical properties such as hydropathy, molecular weight and diversity in secondary structure and thus closely resemble natural proteins. Furthermore, functionally conditioned samples exhibit increased specificity by accurately reproducing active site motifs and patterns of hydrophobicity while preserving sequence diversity. In terms of quality, diversity and novelty metrics, PLAID demonstrates consistent superiority over baseline models, yielding the largest set of unique and designable protein clusters that exhibit realistic structural and functional properties.
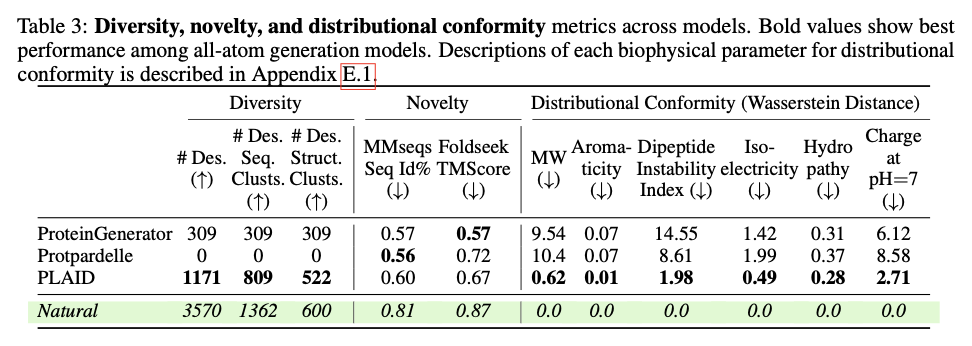
PLAID represents a revolutionary method for protein design that focuses on scalability, data availability, and multimodal integration of grand challenges. In this way, it is possible to link sequence and structure generation by applying sequence-based training and latent diffusion to create diverse, accurate and biologically relevant protein designs. This approach can potentially be used for further applications in molecular engineering, therapeutic development and synthetic biology, opening the doors to new solutions to complex biological problems.
Checkout The Paper and GitHub page. All credit for this research goes to the researchers of this project. Also don't forget to follow us Twitter and join ours Telegram channel And LinkedIn GrOup. Don't forget to join our 60k+ ML SubReddit.
🚨 Trending: LG AI Research Releases EXAONE 3.5: Three Open Source Models at the Bilingual Frontier AI Level, Offering Unmatched Guidance and Long Contextual Understanding for Global Leadership in Generative AI Excellence…
Aswin AK is a consulting intern at MarkTechPost. He is pursuing his double degree from the Indian Institute of Technology, Kharagpur. He has a passion for data science and machine learning and brings a strong academic background and practical experience in solving real-world cross-domain challenges.
🧵🧵 [Download] Assessment of Large Language Model Vulnerabilities Report (Funded)